

- How to install rtools in r studio for mac#
- How to install rtools in r studio software#
- How to install rtools in r studio code#
- How to install rtools in r studio series#
- How to install rtools in r studio mac#
However, there is a small minority of packages, like e.g. Why should I care, again? As long as you stick to base R and add-on packages that are available as pre-compiled binaries at CRAN or Bioconductor (and that is the vastly overwhelming majority of packages), you don’t need to care at all. Which is only possible because the whole tool chain for doing this is open source, again. Which is a world of pain, which is why the good people of the R project have made available an installer which does all that very conveniently, as outlined below. That means, if you want to build your own packages from source on that Winbox, you will need to install all these tools in the correct version.
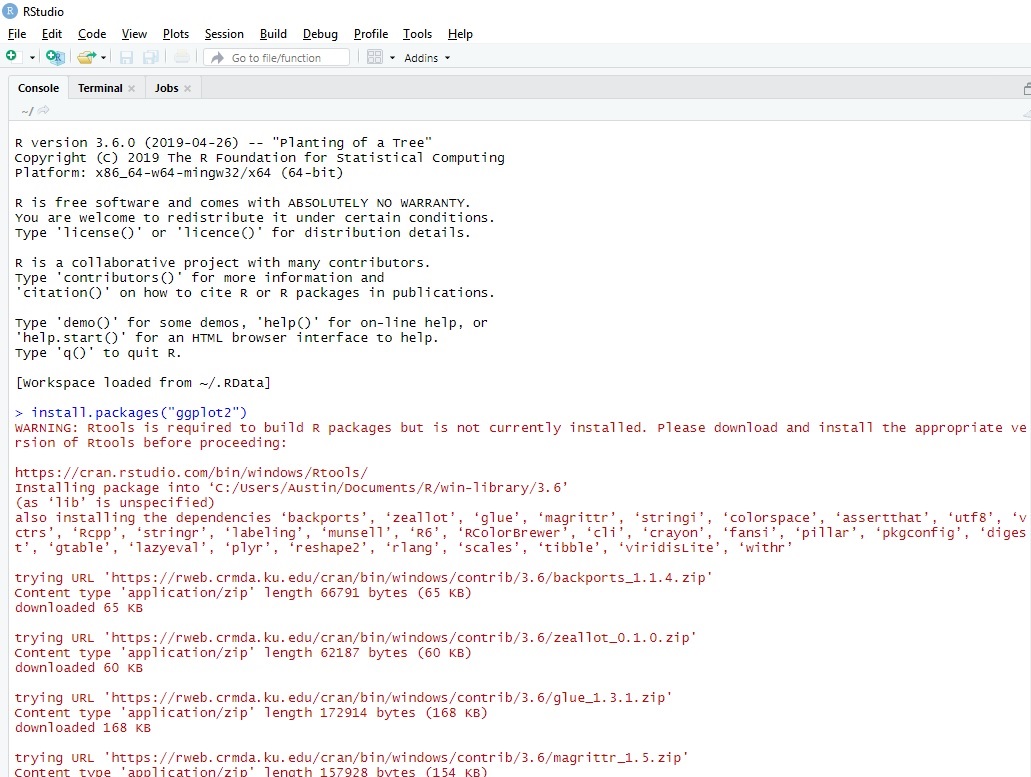
These tools are of course not available on a standard Windows box (or only as cheap commercial knockoffs that will not work). Note that all these little helpers will be available on, say, an out-of-the box Ubuntu installation.
How to install rtools in r studio series#
This in its turn requires a whole series of external tools like make, sed, tar, gzip, a C/C++ compiler etc.
How to install rtools in r studio code#
Note that this is a non-trivial process, involving making different kinds of documentation, checking R code for sanity, possibly compiling C/C++ or Fortran code and so on. What it is Now on Linux (and presumably on most other Unices), R downloads the package source and builds the ready-to-use package that is installed on your system right there on your machine.
How to install rtools in r studio mac#
So what really happens when you install the package on Mac or Windows is that R actually downloads and installs the pre-compiled binary for your operating system, without ever bothering you about the details.

How to install rtools in r studio for mac#
xtable_1.7-0.tgz, the package built for Mac.However, if you visit the home page of the package at CRAN you will find there (currently)

Well, at least it feels that way: install.packages("xtable") would work on any system. RData files, and you install the same packages from CRAN. What counts is that under the hood, everything is pretty much the same: you run the same R commands, you read and write the same. Now even if you don’t feel strongly one way or the other about open source (though you should), the cross-platform aspect is very useful indeed: wherever you go, Windows or Mac or Linux, R will be there for you – just go to CRAN, pick the appropriate installer for your system, and you’re good to go, with virtually no differences.Įxcept, of course, of the obvious ones, in that the GUI may look quite different on a Mac compared to Windows – but that’s all just superficial, mere surface, wrapping the underlying crunchy goodness of the R language slightly differently: the command line is the same, and if you want have GUI consistency, a more elaborate wrapper like JGR or RStudio is going to be your friend.
How to install rtools in r studio software#
Admittedly, these are kind of two things, though you might argue that any sufficiently popular open source software will be ported widely. It will show the documentation of install.packages() in R Studio as shown in below images.Some background One of the great things about R statistical language & environment is that it is open source and cross-platform. To know more about install.packages() function, you can type either ?install.packages() or help(install.packages). Instailing Package By function install.packages() We can also install the packages manually after downloading from R Standard and official website site or from other sources available on Google. We can install a package in R using function install.packages() or following the steps of submenu Install Packages. Some of them are standard packages that come with R installation package, such as - tidyverse, shiny, ggplot2, readxl, devtools, plumber, gdata etc. There are several package (around 5k) available in R. The packages in R is developed by R Studio team as well as individuals. Packages are the collections of the R functions and data sets. Packages are add-ons that can extend R's functionality and perform specific tasks covering a wide range of statistics. In this article, we shall discuss and learn about the packages in R. It is a computing environment where statistical data may be implemented. It is also used in machine learning, data science, research, and many more new fields. R is an important programming language used heavily by statisticians.


 0 kommentar(er)
0 kommentar(er)
